

SLIC SuperPixels
and DBSCAN Clustering
April 2013
This segmentation approach makes use of Achanta et al's SLIC superpixels and the DBSCAN clustering algorithm. Application of the SLIC superpixel algorithm forms an over-segmentation of an image. These superpixels are then processed using the DBSCAN algorithm to form clusters of superpixels to generate the final segmentation. The approach is simple and relatively fast.
The speed of the DBSCAN clustering process is greatly facilitated by forming an adjacency matrix of the regions produced by the super-pixelization process. This constrains the number of distance measurement tests required

Original image. Hallett Cove, South Australia

Superpixels generated by
SLIC
The following code segments the image into 3000 superpixels using a weighting factor of 10 relating spatial distances to colour distances, resulting superpixels of area less than 10 pixels are eliminated, and superpixel attributes are computed from the median colour values. Note I prefer to use a relative low weighting of spatial distance to colour distance in forming superpixels. The SLIC algorithm does not enforce continuity of superpixels which means that superpixels can break up unto multiple regions if the pixels within a region are significantly different and the superpixel weighting of spatial distance to colour distance is low. This is not a problem, indeed I consider it an advantage because it means that with a low weighting you reduce the chance of undersegmenting an image and the fidelity of feature boundaries is better preserved. The disjoint superpixels are made distinct by the functions cleanupregions.m or mcleanupregions.m. Note also that the superpixel seed points have been initialised in a hexagonal grid rather than a square one. The idea is that this results in a segmentation that will be nominally 6-connected which hopefully facilitates any subsequent post-processing that seeks to merge superpixels.
>> [l, Am, C] = slic(im, 3000, 10, 1, 'median'); >> show(drawregionboundaries(l, im, [255 255 255]))

Clustering of superpixels using DBSCAN
Here I have chosen a L*a*b* colour difference threshold of 5. Small changes in this value can have a significant effect.
>> lc = spdbscan(l, C, Am, 5); >> show(drawregionboundaries(lc, im, [255 255 255]))
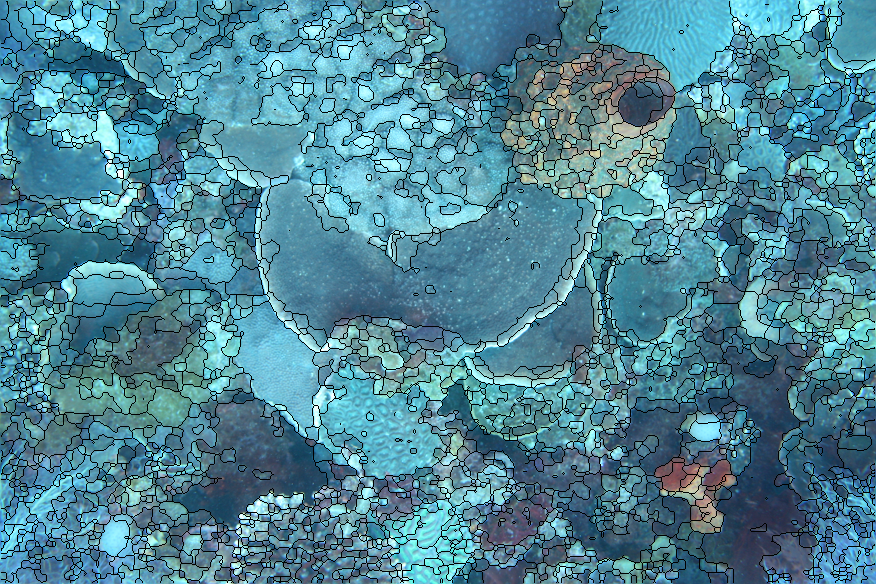
An example of this approach applied to a coral image. As with the Hallett Cove image the result is somewhat oversegmented (and undersegmented in some areas too). Segmentation based just on colour is not sufficient, some texture information is needed too.